Laurens D’Huys

Fluorocode: towards super-resolved mapping of the gut microbiome
Started on September, 2017
Prof. Johan Hofkens
email Laurens D'Huys
Animalia, bacteria and viruses have been part of a delicately balanced system for about as long as they have co-existed, long before the advent of mankind. The gut of our ancient ancestors offered protected, nutrient-rich habitats for bacteria. In return, animals could take an evolutionary short-cut to developing new capabilities by ‘borrowing’ bacterial genes. As gut bacteria assist with the processing of e.g. host-indigestible carbohydrates or the production of vitamins, they will exert significant influence on the host immune system through intricate networks of host molecular pathways or by preventing colonization of the gut by pathogens. As such, commensal bacteria have been implicated in the function and homeostasis of the immune system. Gut bacteria, in addition to host genetics, constitute a major factor in the etiology of complex disorders. They are implicated in obesity, cardiovascular disease, autoimmune diseases like multiple sclerosis, rheumatoid arthrosis and inflammation conditions such as inflammatory bowel disease (IBD) Crohn’s (CD) or even neurological conditions such as Alzheimer’s (AD).
With this work, an approach for enterotype analysis via enzyme-mediated, super-resolution optical genome mapping (Fluorocode) is presented (Figure 1). This methodology will be applied to obtain a diagnostic platform for studying the role of the bacterial and viral (bacteriophage) enterotype in human health and disease. The long range genomic information unlocked through Fluorocode will allow the envisioned platform to complement widely used NGS approaches by providing a more cost-effective alternative in diagnostic applications where base level resolution is not required. In addition, reducing cost and time expense per experimental run will allow for monitoring microbiome changes over time after administration of microbiome modulators (e.g. probiotics), thus also allowing for theragnostic use of the method.
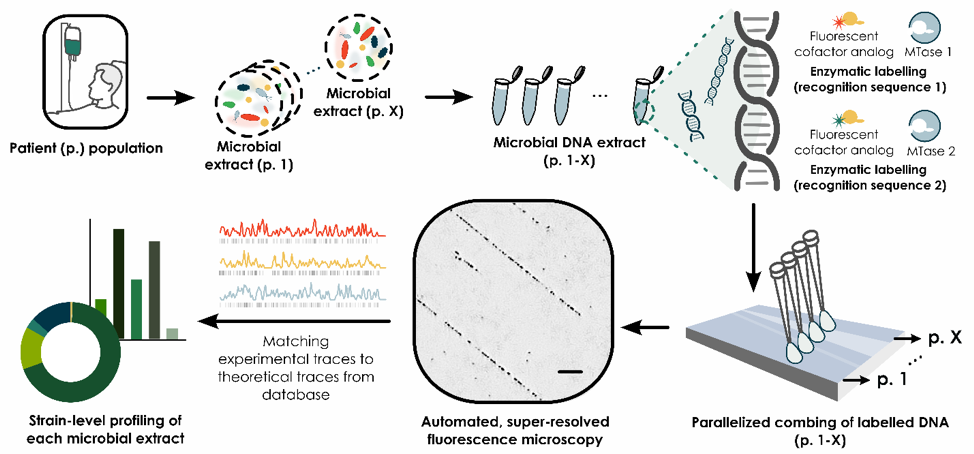
Figure 1. Fluorocode workflow (Mtase = Methyltransferase)
Publications
Arno Bouwens, Jochem Deen, Raffaele Vitale, Laurens D'Huys, Vince Goyvaerts, Adrien Descloux, Doortje Borrenberghs, Kristin Grussmayer, Tomas Lukes, Rafael Camacho, Jia Su, Cyril Ruckebusch, Theo Lasser, Dimitri Van De Ville, Johan Hofkens, Aleksandra Radenovic, Kris Pieter Frans Janssen
Published in bioRxiv, April 2019 (see publication in Journal )
